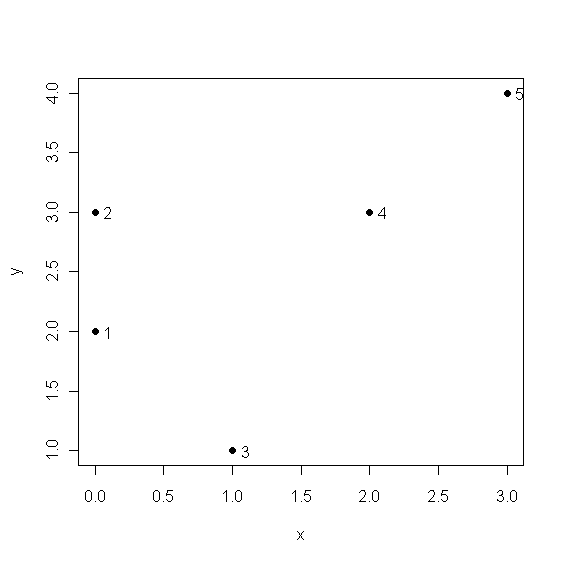Table of Contents
Clustering
8. May 2011
Internal structures of clustering procedures in R
> x <- c(0,0,1,2,3)
> y <- c(2,3,1,3,4)
> U <- cbind(x,y)
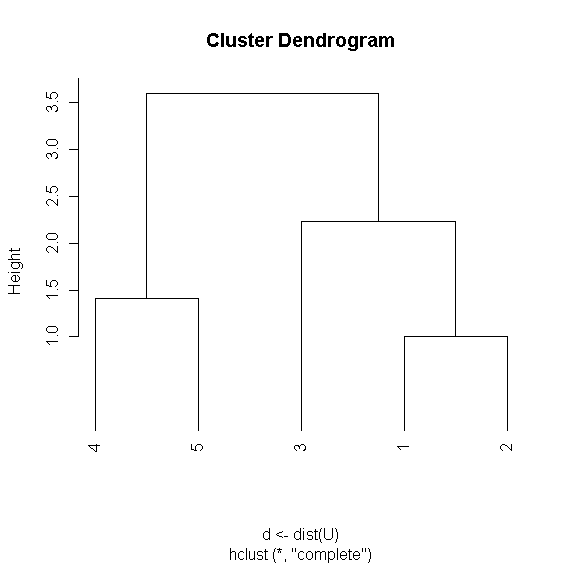
> r <- hclust(d<-dist(U))
> names(r)
[1] "merge" "height" "order" "labels" "method" "call" "dist.method"
> plot(r,hang=-1)
> r$merge
[,1] [,2]
[1,] -1 -2
[2,] -4 -5
[3,] -3 1
[4,] 2 3
> plot(x,y,pch=20,cex=1.5); text(x+0.1,y,1:5)
Procedure for alternative cutting of clusters
varCutree <- function(R,var,vmin,vmax){
mark <- function(t,c){
if(t<0) part[-t] <<- c else {mark(R$merge[t,1],c); mark(R$merge[t,2],c)}
}
n <- length(var); part <- rep(NA,n); nclust <- 0
value <- cbind(var,0)
for(i in 1:(n-1)){
j <- R$merge[i,1]; a <- ifelse(j<0,value[-j,1],value[j,2])
j <- R$merge[i,2]; b <- ifelse(j<0,value[-j,1],value[j,2])
value[i,2] <- a+b
}
value[n,2] <- 0
for(i in 1:(n-1)){
if(value[i,2]<=vmax) next
l <- R$merge[i,1]; a <- ifelse(l<0,value[-l,1],value[l,2])
r <- R$merge[i,2]; b <- ifelse(r<0,value[-r,1],value[r,2])
if(min(a,b)>vmax) next
if(a<=vmax) if(a>=vmin) {nclust <- nclust+1; mark(l,nclust)} else mark(l,0)
if(b<=vmax) if(b>=vmin) {nclust <- nclust+1; mark(r,nclust)} else mark(r,0)
}
return(list(part=part,value=value))
}
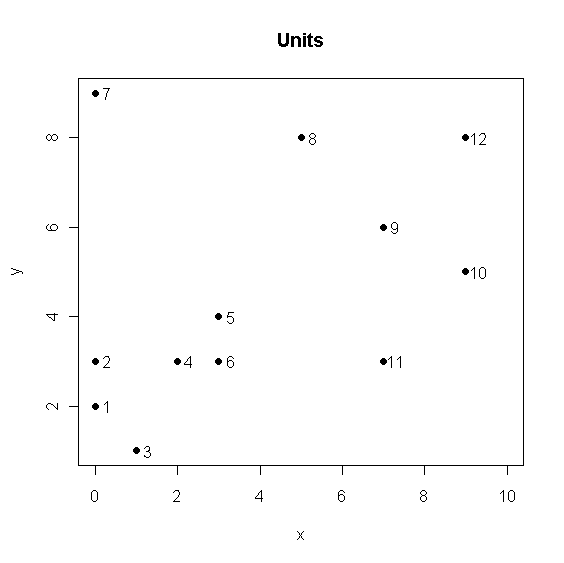
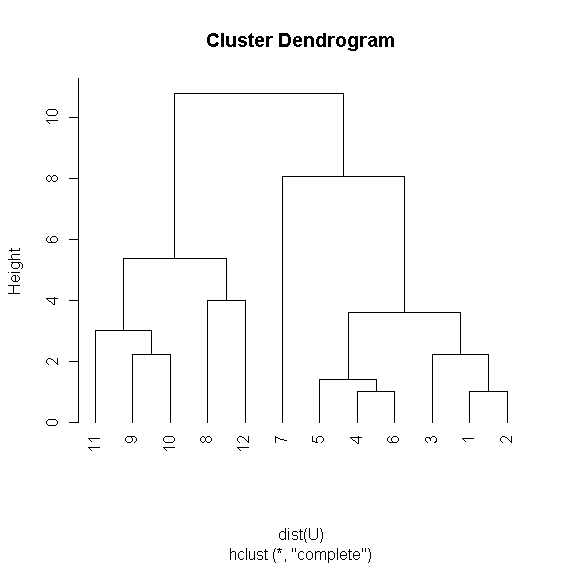
> x <- c(0,0,1,2,3,3,0,5,7,9,7,9) > y <- c(2,3,1,3,4,3,9,8,6,5,3,8) > U <- cbind(x,y) > plot(x,y,pch=20,cex=1.5,xlim=c(0,10),main="Units"); text(x+0.3,y,1:12) > r <- hclust(dist(U)) > plot(r,hang=-1) > rr <- varCutree(r,rep(1,12),2,5) > rr$part [1] 2 2 2 1 1 1 0 3 3 3 3 3 > table(rr$part) 0 1 2 3 1 3 3 5 > (rc <- cutree(r,k=4)) [1] 1 1 1 1 1 1 2 3 4 4 4 3 > table(rc) 1 2 3 4 6 1 2 3
Improved version of procedure for incomplete clusterings
varCutree <- function(R,var,vmin,vmax){
mark <- function(t,c){
if(t<0) part[-t] <<- c else {mark(R$merge[t,1],c); mark(R$merge[t,2],c)}
}
n <- length(var); part <- rep(999999,n); nclust <- 0
value <- cbind(var,rep(0,n))
for(i in 1:(n-1)){
j <- R$merge[i,1]; if(j==0) next
a <- ifelse(j<0,value[-j,1],value[j,2])
j <- R$merge[i,2]; if(j==0) next
b <- ifelse(j<0,value[-j,1],value[j,2])
value[i,2] <- a+b
}
value[n,2] <- 0
for(i in 1:(n-1)){
if(value[i,2]<=vmax) next
l <- R$merge[i,1]; r <- R$merge[i,2]
if(l==0) a <- 0 else a <- ifelse(l<0,value[-l,1],value[l,2])
if(r==0) b <- 0 else b <- ifelse(r<0,value[-r,1],value[r,2])
if(min(a,b)>vmax) next
if(a<=vmax) if(a>=vmin) {nclust <- nclust+1; mark(l,nclust)} else mark(l,0)
if(b<=vmax) if(b>=vmin) {nclust <- nclust+1; mark(r,nclust)} else mark(r,0)
}
return(list(part=part,value=value))
}
Transforming Pajek's hierarchical clusterings into R's
tree <- as.integer(read.csv("./cluster/AvgTol.clu",header=FALSE,skip=1)$V1)
N <- length(tree); n <- (N+1) %/% 2
merge <- matrix(0,nrow=(n-1),ncol=2)
for(i in 1:n) if(tree[i]>0){
k <- tree[i]-n
if(merge[k,1]==0) merge[k,1] <- -i else merge[k,2] <- -i
}
for(i in (n+1):N) if(tree[i]>0){
k <- tree[i]-n; j <- i-n
if(merge[k,1]==0) merge[k,1] <- j else merge[k,2] <- j
}
> RC <- list(merge=merge,n=n)
> rC <- varCutree(RC,rep(1,3110),10,800)
> table(rC$part)
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21
729 794 600 130 97 17 13 10 337 40 20 12 25 19 11 32 116 42 10 18 16 14
> out <- file("CRc.clu","w"); cat("*vertices 3110",rC$part,sep="\n",file=out); close(out)
Derived dendrograms
rankTree <- function(R){
nm <- length(R$height)
rank <- rep(0,nm)
for (i in 1:nm){
j <- R$merge[i,1]; a <- ifelse(j<0,0,rank[j])
j <- R$merge[i,2]; b <- ifelse(j<0,0,rank[j])
rank[i] <- 1+max(a,b)
}
return(rank)
}
countTree <- function(R){
nm <- length(R$height)
count <- rep(1,nm)
for (i in 1:nm){
j <- R$merge[i,1]; a <- ifelse(j<0,1,count[j])
j <- R$merge[i,2]; b <- ifelse(j<0,1,count[j])
count[i] <- a+b
}
return(count)
}
errorTree <- function(R){
nm <- length(R$height)
error <- rep(0,nm)
for (i in 1:nm){
j <- R$merge[i,1]; a <- ifelse(j<0,0,error[j])
j <- R$merge[i,2]; b <- ifelse(j<0,0,error[j])
error[i] <- a+b+R$height[i]
}
return(error)
}
> T = matrix( c( 1, 3, 2, 2, 3, 3, 3, 4, 4, 1, 4, 12, 6, 15, 8, 12,
10, 9, 11, 13, 12, 4, 14, 2, 14, 11, 15, 4, 16, 2 ), ncol=2, byrow=TRUE)
> rownames(T) <- letters[1:nrow(T)]; colnames(T) <- c('X','Y')
> plot(T,main='Example 1'); text(T[,1]+0.3,T[,2]-0.4,rownames(T))
> D <- dist(T)
> t <- hclust(D,method="ward")
> plot(t,hang=-1,cex=0.4,main="Example 1/Ward")
> H <- t$height; N <- rankTree(t); t$height <- N
> plot(t,hang=-1,cex=0.4,main="Example 1/Ward/rank")
> t$height <- H
> E <- errorTree(t); t$height <- E
> plot(t,hang=-1,cex=0.4,main="Example 1/Ward/P")
> t$height <- log(E)
> plot(t,hang=-1,cex=0.4,main="Example 1/Ward/log P")
> t$height <- sqrt(E)
> plot(t,hang=-1,cex=0.4,main="Example 1/Ward/sqrt P")
All three procedures in one
derivedTree <- function(R,type='rank'){
if (type == 'total') { c <- 0; ex <- expression(a+b+R$height[i]) }
else if (type == 'count') { c <- 1; ex <- expression(a+b) }
else { c <- 0; ex <- expression(1+max(a,b)) }
nm <- length(R$height)
h <- rep(c,nm)
for (i in 1:nm){
j <- R$merge[i,1]; a <- ifelse(j<0,c,h[j])
j <- R$merge[i,2]; b <- ifelse(j<0,c,h[j])
h[i] <- eval(ex)
}
return(h)
}
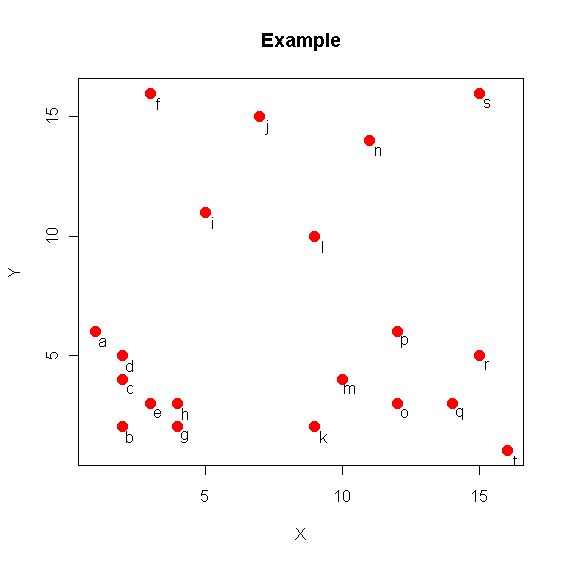
Example
> T = matrix( c( 0, 5, 1, 1, 1, 3, 1, 4, 2, 2, 2, 15,
+ 3, 1, 3, 2, 4, 10, 6, 14, 8, 1, 8, 9, 9, 3,
+ 10, 13, 11, 2, 11, 5, 13, 2, 14, 4, 14, 15, 15, 0 ),
+ ncol=2, byrow=TRUE)
> T <- T+1
> rownames(T) <- letters[1:nrow(T)]; colnames(T) <- c('X','Y')
> plot(T,main='Example',col="red",pch=16,cex=1.5)
> text(T[,1]+0.3,T[,2]-0.4,rownames(T))
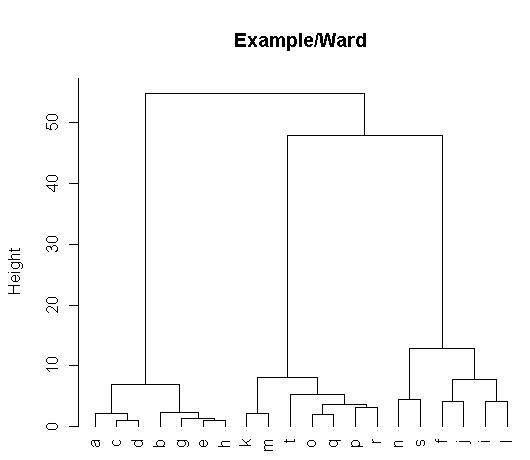
> D <- dist(T)
> t <- hclust(D,method="ward")
> plot(t,hang=-1,cex=1.0,main="Example/Ward")
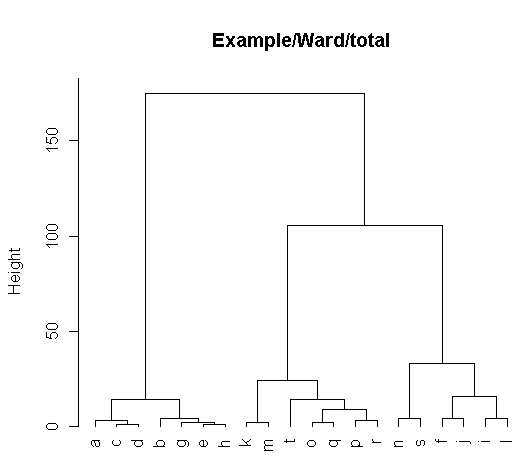
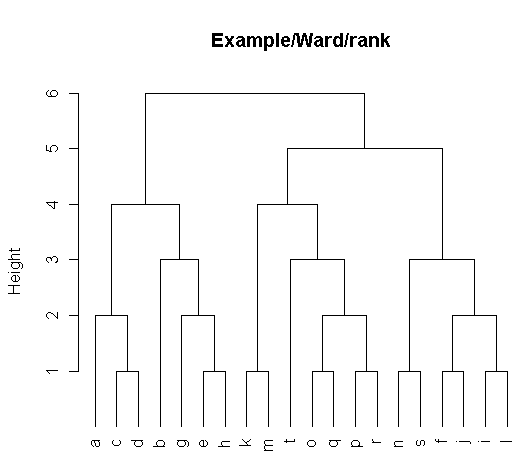
> H <- t$height; t$height <- derivedTree(t,'count')
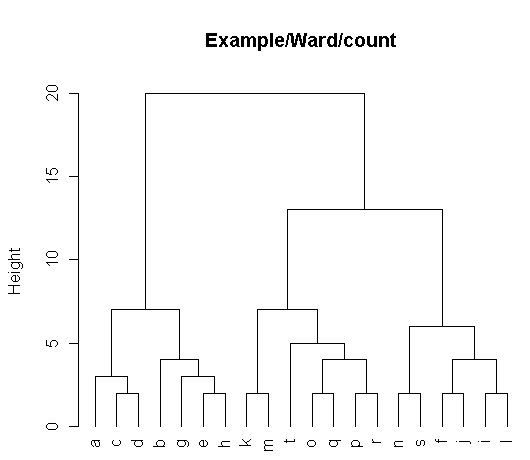
> plot(t,hang=-1,cex=1.0,main="Example/Ward/count")
> t$height <- H; t$height <- derivedTree(t,'rank')
> plot(t,hang=-1,cex=1.0,main="Example/Ward/rank")
> t$height <- H; t$height <- derivedTree(t,'total')
> plot(t,hang=-1,cex=1.0,main="Example/Ward/total")
Except where otherwise noted, content on this wiki is licensed under the following license: CC Attribution-Noncommercial-Share Alike 3.0 Unported