Food SR25
The food data ara available at USDA's National Nutrient Database for Standard Reference. The last (2012) version is SR25.
Reading the data
I selected the abbreviated data, downloaded them and renamed the file abbrev.txt to
abbrev25.txt. Opening the file in text-editor I saw that the column 2 contains the names of foods. I tried to read the data
> setwd("C:/Users/Batagelj/work/clamix/clamix.R")
> a <- read.csv("./sr25/abbrev25.txt",header=FALSE,sep="^",dec=".",quote="~",row.names=2,
+ stringsAsFactors=FALSE)
Error in read.table(file = file, header = header, sep = sep, quote = quote, :
duplicate 'row.names' are not allowed
I decided to add to the end of duplicated names a string ”(1)”
> a <- read.csv("./sr25/abbrev25.txt",header=FALSE,sep="^",dec=".",quote="~",stringsAsFactors=FALSE)
> k <- which(duplicated(a$V2))
> a$V2[k]
[1] "BABYFOOD,MEAT,BF,STR"
[2] "OIL,INDUSTRIAL,PALM KERNEL (HYDROGENATED),CONFECTION FAT"
[3] "BEEF,CHUCK,UNDER BLADE CNTR STEAK,BNLESS,DENVER CUT,LN,0\" FA"
[4] "CAMPBELL'S SEL MICROWAVEABLE BOWLS,HEA"
[5] "POPCORN,OIL-POPPED,LOFAT"
> a$V2[k] <- paste(a$V2[k],"(1)",sep="")
> a$V2[k]
[1] "BABYFOOD,MEAT,BF,STR(1)"
[2] "OIL,INDUSTRIAL,PALM KERNEL (HYDROGENATED),CONFECTION FAT(1)"
[3] "BEEF,CHUCK,UNDER BLADE CNTR STEAK,BNLESS,DENVER CUT,LN,0\" FA(1)"
[4] "CAMPBELL'S SEL MICROWAVEABLE BOWLS,HEA(1)"
[5] "POPCORN,OIL-POPPED,LOFAT(1)"
> rownames(a) <- a$V2
> rownames(a)[1:10]
[1] "SOUP,BF BROTH OR BOUILLON,PDR,DRY" "SOUP,BEEF BROTH,CUBED,DRY"
[3] "SOUP,BF NOODLE,DRY,MIX" "SOUP,CHICK BROTH OR BOUILLON,DRY"
[5] "SOUP,CHICK BROTH CUBES,DRY" "CAMPBELL'S RED & WHITE,CHICK NOODLEO'S SOUP,COND"
[7] "CAMPBELL'S RED & WHITE,CHICK VEG SOUP,COND" "CAMPBELL'S RED & WHITE,BF BROTH,COND"
[9] "SOUP,ONION,DRY,MIX" "CAMPBELLS RED & WHITE,BF CONSOMME,COND"
From the documentation SR25_doc.pdf, pages 37-39 I obtained the list of columns / variables
n Field name Type Description 1 NDB_No. A 5* 5-digit Nutrient Databank number. 2 Shrt_Desc A 60 60-character abbreviated description of food item 3 Water N 10.2 Water (g/100 g) 4 Energ_Kcal N 10 Food energy (kcal/100 g) 5 Protein N 10.2 Protein (g/100 g) 6 Lipid_Tot N 10.2 Total lipid (fat)(g/100 g) 7 Ash N 10.2 Ash (g/100 g) 8 Carbohydrt N 10.2 Carbohydrate, by difference (g/100 g) 9 Fiber_TD N 10.1 Total dietary fiber (g/100 g) 10 Sugar_Tot N 10.2 Total sugars (g/100 g) 11 Calcium N 10 Calcium (mg/100 g) 12 Iron N 10.2 Iron (mg/100 g) 13 Magnesium N 10 Magnesium (mg/100 g) 14 Phosphorus N 10 Phosphorus (mg/100 g) 15 Potassium N 10 Potassium (mg/100 g) 16 Sodium N 10 Sodium (mg/100 g) 17 Zinc N 10.2 Zinc (mg/100 g) 18 Copper N 10.3 Copper (mg/100 g) 19 Manganese N 10.3 Manganese (mg/100 g) 20 Selenium N 10.1 Selenium (µg/100 g) 21 Vit_C N 10.1 Vitamin C (mg/100 g) 22 Thiamin N 10.3 Thiamin (mg/100 g) 23 Riboflavin N 10.3 Riboflavin (mg/100 g) 24 Niacin N 10.3 Niacin (mg/100 g) 25 Panto_acid N 10.3 Pantothenic acid (mg/100 g) 26 Vit_B6 N 10.3 Vitamin B6 (mg/100 g) 27 Folate_Tot N 10 Folate, total (µg/100 g) 28 Folic_acid N 10 Folic acid (µg/100 g) 29 Food_Folate N 10 Food folate (µg/100 g) 30 Folate_DFE N 10 Folate (µg dietary folate equivalents/100 g) 31 Choline_Tot N 10 Choline, total (mg/100 g) 32 Vit_B12 N 10.2 Vitamin B12 (µg/100 g) 33 Vit_A_IU N 10 Vitamin A (IU/100 g) 34 Vit_A_RAE N 10 Vitamin A (µg retinol activity equivalents/100g) 35 Retinol N 10 Retinol (µg/100 g) 36 Alpha_Carot N 10 Alpha-carotene (µg/100 g) 37 Beta_Carot N 10 Beta-carotene (µg/100 g) 38 Beta_Crypt N 10 Beta-cryptoxanthin (µg/100 g) 39 Lycopene N 10 Lycopene (µg/100 g) 40 Lut+Zea N 10 Lutein+zeazanthin (µg/100 g) 41 Vit_E N 10.2 Vitamin E (alpha-tocopherol) (mg/100 g) 42 Vit_D_mcg N 10.1 Vitamin D (µg/100 g) 43 Vit_D_IU N 10 Vitamin D (IU/100 g) 44 Vit_K N 10.1 Vitamin K (phylloquinone) (µg/100 g) 45 FA_Sat N 10.3 Saturated fatty acid (g/100 g) 46 FA_Mono N 10.3 Monounsaturated fatty acids (g/100 g) 47 FA_Poly N 10.3 Polyunsaturated fatty acids (g/100 g) 48 Cholestrl N 10.3 Cholesterol (mg/100 g) 49 GmWt_1 N 9.2 First household weight for this item from the Weight file.‡ 50 GmWt_Desc1 A 120 Description of household weight number 1. 51 GmWt_2 N 9.2 Second household weight for this item from the Weight file.‡ 52 GmWt_Desc2 A 120 Description of household weight number 2. 53 Refuse_Pct N 2 Percent refuse.
From this list we get the variable names
> names(a)[1:10] [1] "V1" "V2" "V3" "V4" "V5" "V6" "V7" "V8" "V9" "V10" > vn <- c( + "NDB_No." , "Shrt_Desc" , "Water" , "Energ_Kcal", "Protein" , "Lipid_Tot" , + "Ash" , "Carbohydrt", "Fiber_TD" , "Sugar_Tot" , "Calcium" , "Iron" , + "Magnesium" , "Phosphorus", "Potassium" , "Sodium" , "Zinc" , "Copper" , + "Manganese" , "Selenium" , "Vit_C" , "Thiamin" , "Riboflavin" , "Niacin" , + "Panto_acid" , "Vit_B6" , "Folate_Tot", "Folic_acid", "Food_Folate", "Folate_DFE" , + "Choline_Tot", "Vit_B12" , "Vit_A_IU" , "Vit_A_RAE" , "Retinol" , "Alpha_Carot", + "Beta_Carot" , "Beta_Crypt", "Lycopene" , "Lut+Zea" , "Vit_E" , "Vit_D_mcg" , + "Vit_D_IU" , "Vit_K" , "FA_Sat" , "FA_Mono" , "FA_Poly" , "Cholestrl" , + "GmWt_1" , "GmWt_Desc1", "GmWt_2" , "GmWt_Desc2", "Refuse_Pct" ) > a <- a[,-2] > names(a) <- vn[-2] > names(a) [1] "NDB_No." "Water" "Energ_Kcal" "Protein" "Lipid_Tot" "Ash" [7] "Carbohydrt" "Fiber_TD" "Sugar_Tot" "Calcium" "Iron" "Magnesium" [13] "Phosphorus" "Potassium" "Sodium" "Zinc" "Copper" "Manganese" [19] "Selenium" "Vit_C" "Thiamin" "Riboflavin" "Niacin" "Panto_acid" [25] "Vit_B6" "Folate_Tot" "Folic_acid" "Food_Folate" "Folate_DFE" "Choline_Tot" [31] "Vit_B12" "Vit_A_IU" "Vit_A_RAE" "Retinol" "Alpha_Carot" "Beta_Carot" [37] "Beta_Crypt" "Lycopene" "Lut+Zea" "Vit_E" "Vit_D_mcg" "Vit_D_IU" [43] "Vit_K" "FA_Sat" "FA_Mono" "FA_Poly" "Cholestrl" "GmWt_1" [49] "GmWt_Desc1" "GmWt_2" "GmWt_Desc2" "Refuse_Pct" > dim(a) [1] 8194 52 > food <- a > save(food,file="./sr25/sr25.Rdata")
I removed the second variable (food name) from the data frame and replaced the variable names with the names from the list. I have finally the data in the form of the R data frame. I save it on the file sr25.Rdata .
Encoding the data
I suppose that the encoding rules encFood.R for SR22 can be useful also for SR25. I copied them to the sr25 directory.
encWater <- list( "[0]" = function(x) x<=0, "(0,5.65]" = function(x) x<=5.65, "(5.65,29.5]" = function(x) x<=29.5, "(29.5,53.9)" = function(x) x< 53.9, "[53.9,62.4)" = function(x) x< 62.4, "[62.4,70.75)" = function(x) x< 70.75, "[70.75,78.05]" = function(x) x<=78.05, "(78.05,88)" = function(x) x< 88, "[88,100]" = function(x) x<=100, "NA" = function(x) TRUE ) encEnergKC <- list( "[0]" = function(x) x<=0, "(0,50]" = function(x) x<=50, "(50,104]" = function(x) x<=104, "(104,160]" = function(x) x<=160, "(160,232]" = function(x) x<=232, "(232,312]" = function(x) x<=312, "(312,386]" = function(x) x<=386, "(386,800)" = function(x) x< 800, "[800,905]" = function(x) x<=905, "NA" = function(x) TRUE ) encProtein <- list( "[0]" = function(x) x<=0, "(0,1.5]" = function(x) x<=1.5, "(1.5,4.1]" = function(x) x<=4.1, "(4.1,8.4)" = function(x) x< 8.4, "[8.4,16]" = function(x) x<=16, "(16,23.05)" = function(x) x< 23.05, "[23.05,37]" = function(x) x<=37, "(37,75)" = function(x) x< 75, "[75,90]" = function(x) x<=90, "NA" = function(x) TRUE ) encTotLipi <- list( "[0]" = function(x) x<=0, "(0,0.3]" = function(x) x<=0.3, "(0.3,1.3)" = function(x) x< 1.3, "[1.3,3.8)" = function(x) x< 3.8, "[3.8,8)" = function(x) x< 8, "[8,13.7)" = function(x) x< 13.7, "[13.7,23.5)" = function(x) x< 23.5, "[23.5,85)" = function(x) x< 85, "[85,100]" = function(x) x<=100, "NA" = function(x) TRUE ) encAsh <- list( "[0]" = function(x) x<=0, "(0,0.7)" = function(x) x< 0.7, "[0.7,1)" = function(x) x< 1, "[1,1.24)" = function(x) x< 1.24, "[1.24,1.75)" = function(x) x< 1.75, "[1.75,3)" = function(x) x< 3, "[3,45)" = function(x) x< 45, "[45,95)" = function(x) x< 95, "[95,100]" = function(x) x<=100, "NA" = function(x) TRUE ) encCarbohyd <- list( "[0]" = function(x) x<=0, "(0,3.5]" = function(x) x<=3.5, "(3.5,6.85]" = function(x) x<=6.85, "(6.85,11.35]" = function(x) x<=11.35, "(11.35,18.1)" = function(x) x< 18.1, "[18.1,27.8]" = function(x) x<=27.8, "(27.8,55.1]" = function(x) x<=55.1, "(55.1,73]" = function(x) x<=73, "(73,100]" = function(x) x<=100, "NA" = function(x) TRUE ) encFiberTd <- list( "[0]" = function(x) x<=0, "(0,0.8]" = function(x) x<=0.8, "(0.8,1.5]" = function(x) x<=1.5, "(1.5,2.1]" = function(x) x<=2.1, "(2.1,3.1]" = function(x) x<=3.1, "(3.1,5.6]" = function(x) x<=5.6, "(5.6,35]" = function(x) x<=35, "(35,60]" = function(x) x<=60, "(60,90]" = function(x) x<=90, "NA" = function(x) TRUE ) encCalcium <- list( "[0]" = function(x) x<=0, "(0,7]" = function(x) x<=7, "(7,12]" = function(x) x<=12, "(12,20]" = function(x) x<=20, "(20,34]" = function(x) x<=34, "(34,69]" = function(x) x<=69, "(69,159]" = function(x) x<=159, "(159,3000]" = function(x) x<=3000, "(3000,7400]" = function(x) x<=7400, "NA" = function(x) TRUE ) encIron <- list( "[0]" = function(x) x<=0, "(0,0.35]" = function(x) x<=0.35, "(0.35,0.75]" = function(x) x<=0.75, "(0.75,1.2]" = function(x) x<=1.2, "(1.2,1.8]" = function(x) x<=1.8, "(1.8,2.5]" = function(x) x<=2.5, "(2.5,3.85]" = function(x) x<=3.85, "(3.85,80]" = function(x) x<=80, "(80,125]" = function(x) x<=125, "NA" = function(x) TRUE ) encMagnesiu <- list( "[0]" = function(x) x<=0, "(0,9)" = function(x) x< 9, "[9,16)" = function(x) x< 16, "[16,21)" = function(x) x< 21, "[21,25)" = function(x) x< 25, "[25,32)" = function(x) x< 32, "[32,65)" = function(x) x< 65, "[65,600)" = function(x) x< 600, "[600,900]" = function(x) x<=900, "NA" = function(x) TRUE ) encPhosphor <- list( "[0]" = function(x) x<=0, "(0,25)" = function(x) x< 25, "[25,60)" = function(x) x< 60, "[60,112)" = function(x) x< 112, "[112,172)" = function(x) x< 172, "[172,208)" = function(x) x< 208, "[208,268)" = function(x) x< 268, "[268,6000)" = function(x) x< 6000, "[6000,10000]" = function(x) x<=10000, "NA" = function(x) TRUE ) encPotassium <- list( "[0]" = function(x) x<=0, "(0,90)" = function(x) x< 90, "[90,147)" = function(x) x< 147, "[147,211)" = function(x) x< 211, "[211,281)" = function(x) x< 281, "[281,340)" = function(x) x< 340, "[340,426)" = function(x) x< 426, "[426,6000)" = function(x) x< 6000, "[6000,17000]" = function(x) x<=17000, "NA" = function(x) TRUE ) encSodium <- list( "[0]" = function(x) x<=0, "(0,10)" = function(x) x< 10, "[10,50)" = function(x) x< 50, "[50,66)" = function(x) x< 66, "[66,121)" = function(x) x< 121, "[121,351)" = function(x) x< 351, "[351,655)" = function(x) x< 655, "[655,15000)" = function(x) x< 15000, "[15000,30000]" = function(x) x<=30000, "NA" = function(x) TRUE ) encZinc <- list( "[0]" = function(x) x<=0, "(0,0.23)" = function(x) x< 0.23, "[0.23,0.53)" = function(x) x< 0.53, "[0.53,1.06)" = function(x) x< 1.06, "[1.06,2.23)" = function(x) x< 2.23, "[2.23,4.12)" = function(x) x< 4.12, "[4.12,20)" = function(x) x< 20, "[20,150)" = function(x) x< 150, "[150,200]" = function(x) x<=200, "NA" = function(x) TRUE ) encCopper <- list( "[0]" = function(x) x<=0, "(0,0.045)" = function(x) x< 0.045, "[0.045,0.071)" = function(x) x< 0.071, "[0.071,0.1)" = function(x) x< 0.1, "[0.1,0.135)" = function(x) x< 0.135, "[0.135,0.23)" = function(x) x< 0.23, "[0.23,0.95)" = function(x) x< 0.95, "[0.95,6.5)" = function(x) x< 6.5, "[6.5,10]" = function(x) x<=10, "NA" = function(x) TRUE ) encManganes <- list( "[0]" = function(x) x<=0, "(0,0.016)" = function(x) x< 0.016, "[0.016,0.028)" = function(x) x< 0.028, "[0.028,0.118)" = function(x) x< 0.118, "[0.118,0.27)" = function(x) x< 0.27, "[0.27,0.71)" = function(x) x< 0.71, "[0.71,10)" = function(x) x< 10, "[10,70)" = function(x) x< 70, "[70,80]" = function(x) x<=80, "NA" = function(x) TRUE ) encSelenium <- list( "[0]" = function(x) x<=0, "(0,0.8)" = function(x) x< 0.8, "[0.8,3)" = function(x) x< 3, "[3,11]" = function(x) x<=11, "(11,20)" = function(x) x< 20, "[20,28.5)" = function(x) x< 28.5, "[28.5,160)" = function(x) x< 160, "[160,1500)" = function(x) x< 1500, "[1500,3000]" = function(x) x<=3000, "NA" = function(x) TRUE ) encVitA <- list( "[0]" = function(x) x<=0, "(0,15]" = function(x) x<=15, "(15,52]" = function(x) x<=52, "(52,118]" = function(x) x<=118, "(118,230]" = function(x) x<=230, "(230,560]" = function(x) x<=560, "(560,1800]" = function(x) x<=1800, "(1800,50000]" = function(x) x<=50000, "(50000,100000]"= function(x) x<=100000, "NA" = function(x) TRUE ) encVitE <- list( "[0]" = function(x) x<=0, "(0,0.13)" = function(x) x< 0.13, "[0.13,0.2]" = function(x) x<=0.2, "(0.2,0.3]" = function(x) x<=0.3, "(0.3,0.63]" = function(x) x<=0.63, "(0.63,1.5]" = function(x) x<=1.5, "(1.5,20)" = function(x) x< 20, "[20,190]" = function(x) x<=190, "(190,195]" = function(x) x<=195, "NA" = function(x) TRUE ) encVitC <- list( "[0]" = function(x) x<=0, "(0,0.5)" = function(x) x< 0.5, "[0.5,1]" = function(x) x<=1, "(1,2.5)" = function(x) x< 2.5, "[2.5,6)" = function(x) x< 6, "[6,13.5)" = function(x) x< 13.5, "[13.5,34)" = function(x) x< 34, "[34,1500)" = function(x) x< 1500, "[1500,2400]" = function(x) x<=2400, "NA" = function(x) TRUE ) encThiamin <- list( "[0]" = function(x) x<=0, "(0,0.025]" = function(x) x<=0.025, "(0.025,0.05]" = function(x) x<=0.05, "(0.05,0.08)" = function(x) x<=0.08, "[0.08,0.105]" = function(x) x<=0.105, "(0.105,0.19)" = function(x) x< 0.19, "[0.19,0.42)" = function(x) x< 0.42, "[0.42,8)" = function(x) x< 8, "[8,15]" = function(x) x<=15, "NA" = function(x) TRUE ) encRibolfla <- list( "[0]" = function(x) x<=0, "(0,0.036]" = function(x) x<=0.036, "(0.036,0.076]" = function(x) x<=0.076, "(0.076,0.14)" = function(x) x< 0.14, "[0.14,0.192)" = function(x) x< 0.192, "[0.192,0.25]" = function(x) x<=0.25, "(0.25,0.36]" = function(x) x<=0.36, "(0.36,5]" = function(x) x<=5, "(5,7]" = function(x) x<=7, "NA" = function(x) TRUE ) encNiacin <- list( "[0]" = function(x) x<=0, "(0,0.35]" = function(x) x<=0.35, "(0.35,0.93)" = function(x) x< 0.93, "[0.93,2.26)" = function(x) x< 2.26, "[2.26,3.75]" = function(x) x<=3.75, "(3.75,5.41]" = function(x) x<=5.41, "(5.41,25]" = function(x) x<=25, "(25,60)" = function(x) x< 60, "[60,80]" = function(x) x<=80, "NA" = function(x) TRUE ) encPantoAc <- list( "[0]" = function(x) x<=0, "(0,0.125)" = function(x) x< 0.125, "[0.125,0.27)" = function(x) x< 0.27, "[0.27,0.36]" = function(x) x<=0.36, "(0.36,0.53]" = function(x) x<=0.53, "(0.53,0.83)" = function(x) x< 0.83, "[0.83,15)" = function(x) x< 15, "[15,30)" = function(x) x< 30, "[30,40]" = function(x) x<=40, "NA" = function(x) TRUE ) encVitB6 <- list( "[0]" = function(x) x<=0, "(0,0.037]" = function(x) x<=0.037, "(0.037,0.07]" = function(x) x<=0.07, "(0.07,0.12]" = function(x) x<=0.12, "(0.12,0.215]" = function(x) x<=0.215, "(0.215,0.33]" = function(x) x<=0.33, "(0.33,0.43]" = function(x) x<=0.43, "(0.43,5)" = function(x) x< 5, "[5,8]" = function(x) x<=8, "NA" = function(x) TRUE ) encFolate <- list( "[0]" = function(x) x<=0, "(0,5)" = function(x) x< 5, "[5,8)" = function(x) x< 8, "[8,12)" = function(x) x< 12, "[12,23)" = function(x) x< 23, "[23,48)" = function(x) x< 48, "[48,115)" = function(x) x< 115, "[115,1000)" = function(x) x< 1000, "[1000,2350]" = function(x) x<=2350, "NA" = function(x) TRUE ) encVitB12 <- list( "[0]" = function(x) x<=0, "(0,0.12]" = function(x) x<=0.12, "(0.12,0.3]" = function(x) x<=0.3, "(0.3,0.6]" = function(x) x<=0.6, "(0.6,1.4]" = function(x) x<=1.4, "(1.4,2.47]" = function(x) x<=2.47, "(2.47,3)" = function(x) x< 3, "[3,60)" = function(x) x< 60, "[60,120]" = function(x) x<=120, "NA" = function(x) TRUE ) encFaSat <- list( "[0]" = function(x) x<=0, "(0,0.054)" = function(x) x< 0.054, "[0.054,0.29)" = function(x) x< 0.29, "[0.29,1.08)" = function(x) x< 1.08, "[1.08,2.4)" = function(x) x< 2.4, "[2.4,4.3]" = function(x) x<=4.3, "(4.3,7.9]" = function(x) x<=7.9, "(7.9,80)" = function(x) x< 80, "[80,100]" = function(x) x<=100, "NA" = function(x) TRUE ) encFaMono <- list( "[0]" = function(x) x<=0, "(0,0.035]" = function(x) x<=0.035, "(0.035,0.3]" = function(x) x<=0.3, "(0.3,1.25)" = function(x) x< 1.25, "[1.25,3]" = function(x) x<=3, "(3,5.6)" = function(x) x< 5.6, "[5.6,9.5)" = function(x) x< 9.5, "[9.5,65)" = function(x) x< 65, "[65,85]" = function(x) x<=85, "NA" = function(x) TRUE ) encFaPoly <- list( "[0]" = function(x) x<=0, "(0,0.115]" = function(x) x<=0.115, "(0.115,0.335]" = function(x) x<=0.335, "(0.335,0.685]" = function(x) x<=0.685, "(0.685,1.23)" = function(x) x< 1.23, "[1.23,2.73)" = function(x) x< 2.73, "[2.73,40)" = function(x) x< 40, "[40,60)" = function(x) x< 60, "[60,75]" = function(x) x<=75, "NA" = function(x) TRUE ) encCholestr <- list( "[0]" = function(x) x<=0, "(0,12]" = function(x) x<=12, "(12,45]" = function(x) x<=45, "(45,66]" = function(x) x<=66, "(66,80]" = function(x) x<=80, "(80,94]" = function(x) x<=94, "(94,550]" = function(x) x<=550, "(550,1900]" = function(x) x<=1900, "(1900,3100]" = function(x) x<=3100, "NA" = function(x) TRUE )
May be we can include into analysis some additional variables. Then we need to add also the encoding rules for them.
Here is the list of variables and corresponding rules
1 NDB_No.
2 Water encWater
3 Energ_Kcal encEnergKC
4 Protein encProtein
5 Lipid_Tot encTotLipi
6 Ash encAsh
7 Carbohydrt encCarbohyd
8 Fiber_TD encFiberTd
9 Sugar_Tot
10 Calcium encCalcium
11 Iron encIron
12 Magnesium encMagnesiu
13 Phosphorus encPhosphor
14 Potassium encPotassium
15 Sodium encSodium
16 Zinc encZinc
17 Copper encCopper
18 Manganese encManganes
19 Selenium encSelenium
20 Vit_C encVitC
21 Thiamin encThiamin
22 Riboflavin encRibolfla
23 Niacin encNiacin
24 Panto_acid encPantoAc
25 Vit_B6 encVitB6
26 Folate_Tot
27 Folic_acid
28 Food_Folate
29 Folate_DFE
30 Choline_Tot
31 Vit_B12 encVitB12
32 Vit_A_IU
33 Vit_A_RAE
34 Retinol
35 Alpha_Carot
36 Beta_Carot
37 Beta_Crypt
38 Lycopene
39 Lut+Zea
40 Vit_E encVitE
41 Vit_D_mcg
42 Vit_D_IU
43 Vit_K
44 FA_Sat encFaSat
45 FA_Mono encFaMono
46 FA_Poly encFaPoly
47 Cholestrl encCholestr
48 GmWt_1
49 GmWt_Desc1
50 GmWt_2
51 GmWt_Desc2
52 Refuse_Pct
encVitA
encFolate
Now we can read the data in R, select the variables to be transformed and match them with the corresponding encoding rules - prepare the vectors s and enc.
> setwd("C:/Users/Batagelj/work/clamix/clamix.R")
> load("./sr25/sr25.Rdata")
> (numSO <- nrow(food))
[1] 8194
> source("C:\\Users\\Batagelj\\work\\clamix\\clamix.R\\clamix3.R")
> source("C:\\Users\\Batagelj\\work\\clamix\\clamix.R\\sr25\\encFood.R")
> (s <- c(2:8,10:25,31,40,44:47))
[1] 2 3 4 5 6 7 8 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 31 40
[26] 44 45 46 47
> nVar <- length(s)
> enc <- c("encWater", "encEnergKC", "encProtein", "encTotLipi", "encAsh",
+ "encCarbohyd", "encFiberTd", "encCalcium", "encIron", "encMagnesiu",
+ "encPhosphor", "encPotassium", "encSodium", "encZinc", "encCopper",
+ "encManganes", "encSelenium", "encVitC", "encThiamin", "encRibolfla",
+ "encNiacin", "encPantoAc", "encVitB6", "encVitB12", "encVitE",
+ "encFaSat", "encFaMono", "encFaPoly", "encCholestr" )
>
> for(i in 1:nVar) cat(i,s[i],names(food)[s[i]]," > ",enc[i],'\n')
1 2 Water > encWater
2 3 Energ_Kcal > encEnergKC
3 4 Protein > encProtein
4 5 Lipid_Tot > encTotLipi
5 6 Ash > encAsh
6 7 Carbohydrt > encCarbohyd
7 8 Fiber_TD > encFiberTd
8 10 Calcium > encCalcium
9 11 Iron > encIron
10 12 Magnesium > encMagnesiu
11 13 Phosphorus > encPhosphor
12 14 Potassium > encPotassium
13 15 Sodium > encSodium
14 16 Zinc > encZinc
15 17 Copper > encCopper
16 18 Manganese > encManganes
17 19 Selenium > encSelenium
18 20 Vit_C > encVitC
19 21 Thiamin > encThiamin
20 22 Riboflavin > encRibolfla
21 23 Niacin > encNiacin
22 24 Panto_acid > encPantoAc
23 25 Vit_B6 > encVitB6
24 31 Vit_B12 > encVitB12
25 40 Vit_E > encVitE
26 44 FA_Sat > encFaSat
27 45 FA_Mono > encFaMono
28 46 FA_Poly > encFaPoly
29 47 Cholestrl > encCholestr
The variables and rules match.
Now we are ready to encode all selected variables.
foodSO <- vector("list",nVar)
for(i in 1:nVar){
var <- food[[s[[i]]]]
foodSO[[i]] <- sapply(var,function(x) encodeSO(x,get(enc[[i]]),10))
names(foodSO)[[i]] <- names(food)[[s[[i]]]]
}
Let us prepare also the metadata. First the names of categories for each variable.
> nVarP <- nVar+1
> varCats <- vector("list",nVar)
> varCats[[1]] <- names(encWater)
> varCats[[2]] <- names(encEnergKC)
> varCats[[3]] <- names(encProtein)
> varCats[[4]] <- names(encTotLipi)
> varCats[[5]] <- names(encAsh)
> varCats[[6]] <- names(encCarbohyd)
> varCats[[7]] <- names(encFiberTd)
> varCats[[8]] <- names(encCalcium)
> varCats[[9]] <- names(encIron)
> varCats[[10]] <- names(encMagnesiu)
> varCats[[11]] <- names(encPhosphor)
> varCats[[12]] <- names(encPotassium)
> varCats[[13]] <- names(encSodium)
> varCats[[14]] <- names(encZinc)
> varCats[[15]] <- names(encCopper)
> varCats[[16]] <- names(encManganes)
> varCats[[17]] <- names(encSelenium)
> varCats[[18]] <- names(encVitC)
> varCats[[19]] <- names(encThiamin)
> varCats[[20]] <- names(encRibolfla)
> varCats[[21]] <- names(encNiacin)
> varCats[[22]] <- names(encPantoAc)
> varCats[[23]] <- names(encVitB6)
> varCats[[24]] <- names(encVitB12)
> varCats[[25]] <- names(encVitE)
> varCats[[26]] <- names(encFaSat)
> varCats[[27]] <- names(encFaMono)
> varCats[[28]] <- names(encFaPoly)
> varCats[[29]] <- names(encCholestr)
>
> varCats[[15]]
[1] "[0]" "(0,0.045)" "[0.045,0.071)" "[0.071,0.1)"
[5] "[0.1,0.135)" "[0.135,0.23)" "[0.23,0.95)" "[0.95,6.5)"
[9] "[6.5,10]" "NA"
> nCats <- sapply(varCats,length)
> names(varCats) <- names(foodSO)
Let us create the empty symbolic object so and symbolic object namedSO with named components and save the metadata.
> long <- rownames(food) > so <- emptySO(nCats) > namedSO <- so > names(namedSO) <- names(varCats) > for(i in 1:nVar) varCats[[i]] <- c(varCats[[i]],"num") > for(i in 1:nVar) names(namedSO[[i]]) <- varCats[[i]] > save(nVar,nVarP,so,namedSO,numSO,long,varCats,file="./sr25/sr25.meta")
Finally we transform the recoded data into symbolic objects describing single units and save them.
> SOs <- vector("list",numSO)
> for(i in 1:numSO){
+ st <- so
+ for(j in 1:nVar)
+ {st[[j]][foodSO[[j]][[i]]] <- 1; st[[j]][[nCats[[j]]+1]] <- 1}
+ # names(SOs)[[i]] <- long[[i]]
+ SOs[[i]] <- st
+ }
> save(nVar,nVarP,so,numSO,SOs,file="./sr25/sr25.so")
Basic quantities
nVar- number of variablesnVarP = nVar+1long- names of unitsSOs- set of units represented as symbolic objectsnumSO- number of symbolic objects (units)varCats- list with names variables and names of categories for each variablenCats- number of categories for each variableso- empty symbolic objectnamedSO- empty symbolic object with named components
The main idea is that all computations are done on SOs without named components (to reduce the work) and to use names only in the presentation of results.
Automatic encoding
An approach to determine the encoding rules for a numerical variable is to select the number of categories (bins) and then determine the breaks so that each category gets approximately the same number of units.
Water
For example, for a variable water into 10 categories:
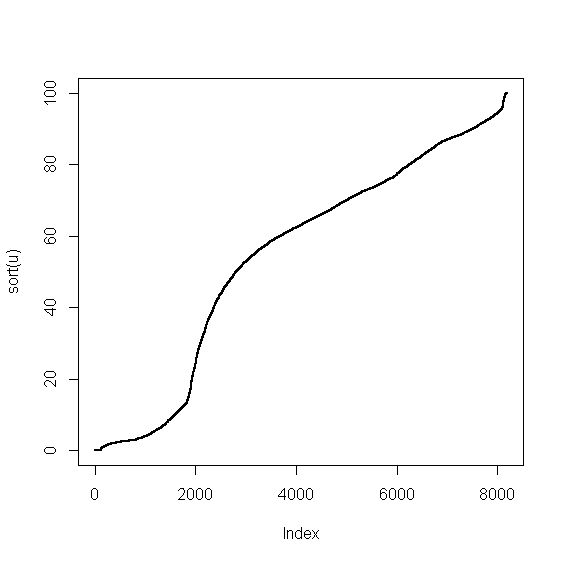
> v <- food$Water > u <- v[!is.na(v)] > plot(sort(u),pch=20,cex=0.5)
The picture doesn't show any special irregularities. We can proceed with encoding
> (brks <- quantile(u, seq(0,1,1/10)))
0% 10% 20% 30% 40% 50% 60% 70% 80% 90%
0.000 3.000 10.504 42.621 56.138 62.980 69.202 74.860 82.836 88.910
100%
100.000
> r <- findInterval(v,brks)
> table(r)
r
1 2 3 4 5 6 7 8 9 10 11
732 906 819 818 818 820 817 820 818 816 4
> r[r==11] <- 10
> (T <- c(as.vector(table(r)),length(r[is.na(r)])))
[1] 732 906 819 818 818 820 817 820 818 820 6
At the end of table we append the number of NA values.
Cholesterol
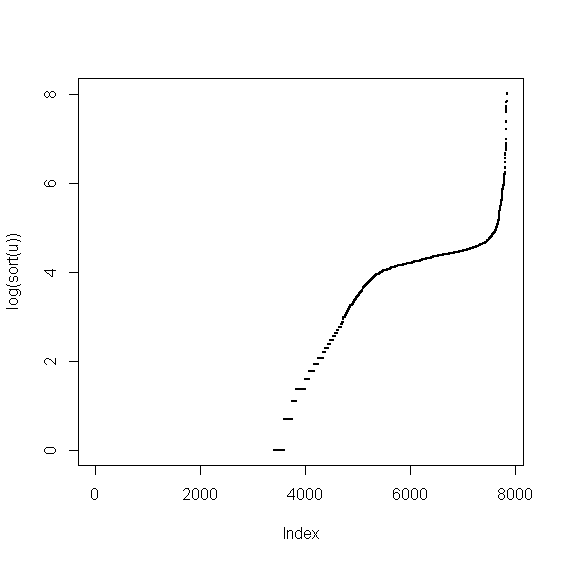
> v <- food$Cholestrl > u <- v[!is.na(v)] > plot(sort(u),pch=20,cex=0.5)
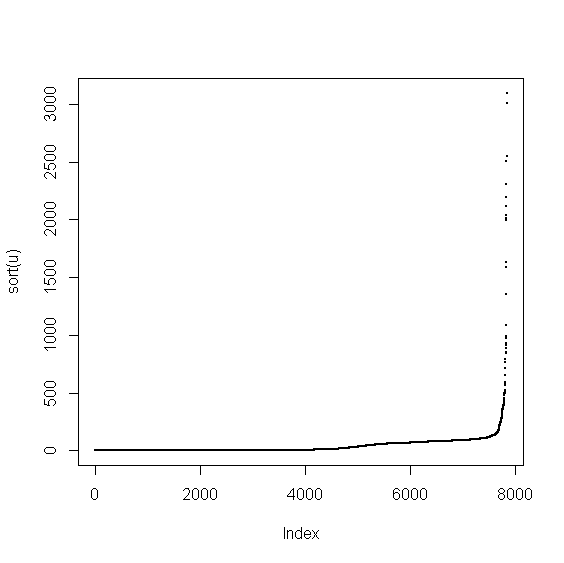 The range of variable's values is large. We'll get a better picture in log-scale
The range of variable's values is large. We'll get a better picture in log-scale
> plot(log(sort(u)),pch=20,cex=0.5)
we see that there are many zero values. We determine the breaks on the positive values and then combine them with zeros and NAs.
> (brks <- quantile(u[u>0], seq(0,1,1/9)))
0% 11.11111% 22.22222% 33.33333% 44.44444% 55.55556% 66.66667% 77.77778%
1 4 10 27 53 66 76 86
88.88889% 100%
102 3100
> r <- findInterval(v,brks)
> r[r==10] <- 9
> (T <- c(as.vector(table(r)),length(r[is.na(r)])))
[1] 3413 415 554 503 486 491 483 494 488 507 360
> a <- c("0",as.character(brks))
> names(T) <- c(paste("[",a[1:10],",",a[2:11],")",sep=""),"NA")
> T
[0,1) [1,4) [4,10) [10,27) [27,53) [53,66)
3413 415 554 503 486 491
[66,76) [76,86) [86,102) [102,3100) NA
483 494 488 507 360
Beta-cryptoxanthin
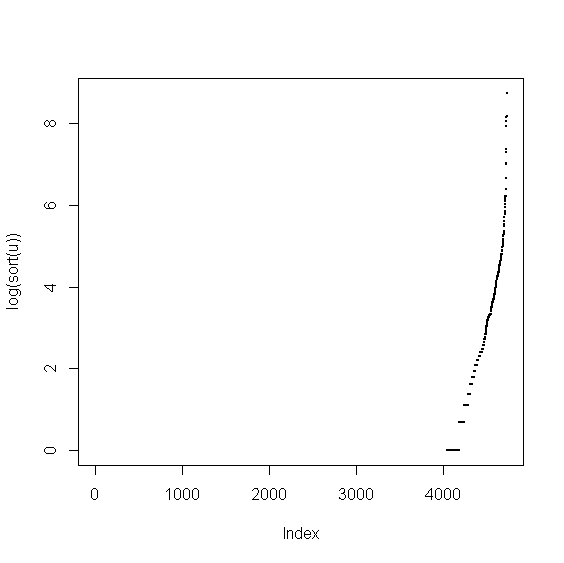
> v <- food$Beta_Crypt > u <- v[!is.na(v)] > plot(log(sort(u)),pch=20,cex=0.5)
From the picture we see that there are many zero and many NA values. We proceed similary as in the case of Cholesterol.
> (brks <- quantile(u[u>0], seq(0,1,1/10)))
0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100%
1.0 1.0 1.0 3.0 5.0 8.0 12.0 26.0 47.0 108.6 6252.0
> bs <- c(1,2,as.vector(brks)[4:11])
> bs
[1] 1.0 2.0 3.0 5.0 8.0 12.0 26.0 47.0 108.6 6252.0
> r <- findInterval(v,bs)
> table(r)
r
0 1 2 3 4 5 6 7 8 9 10
4039 144 61 62 65 80 67 73 70 69 1
> r[r==10] <- 9
> table(r)
r
0 1 2 3 4 5 6 7 8 9
4039 144 61 62 65 80 67 73 70 70
> (T <- c(as.vector(table(r)),length(r[is.na(r)])))
[1] 4039 144 61 62 65 80 67 73 70 70 3463
> names(T) <- c("<1","<2",paste("<",as.character(brks)[4:10],sep=""),
+ paste("<=",as.character(brks)[11],sep=""),"NA")
> T
<1 <2 <3 <5 <8 <12 <26 <47 <108.6 <=6252 NA
4039 144 61 62 65 80 67 73 70 70 3463
> length(v[!is.na(v)&(12<=v)&(v<26)])
[1] 67
> a <- c("0","1","2",as.character(brks)[4:11])
> names(T) <- c(paste("[",a[1:9],",",a[2:10],")",sep=""),
+ paste("[",a[10],",",a[11],"]",sep=""))
> T
[0,1) [1,2) [2,3) [3,5) [5,8) [8,12)
4039 144 61 62 65 80
[12,26) [26,47) [47,108.6) [108.6,6252] <NA>
67 73 70 70 3463
Folic acid
> v <- food$Folic_acid
> u <- v[!is.na(v)]
> plot(log(sort(u)),pch=20,cex=0.5)
> (brks <- quantile(u[u>0], seq(0,1,1/9)))
0% 11.11111% 22.22222% 33.33333% 44.44444% 55.55556% 66.66667% 77.77778%
1.0000 9.0000 17.0000 25.0000 38.0000 54.0000 77.0000 140.0000
88.88889% 100%
328.6667 3269.0000
> r <- findInterval(v,brks)
> table(r)
r
0 1 2 3 4 5 6 7 8 9 10
5273 112 133 120 127 126 126 124 125 124 1
> r[r==10] <- 9
> a <- c("0",as.character(brks))
> (T <- c(as.vector(table(r)),length(r[is.na(r)])))
[1] 5273 112 133 120 127 126 126 124 125 125 1803
> names(T) <- c(paste("[",a[1:10],",",a[2:11],")",sep=""),"NA")
> T
[0,1) [1,9) [9,17)
5273 112 133
[17,25) [25,38) [38,54)
120 127 126
[54,77) [77,140) [140,328.666666666666)
126 124 125
[328.666666666666,3269) NA
125 1803
Analysis
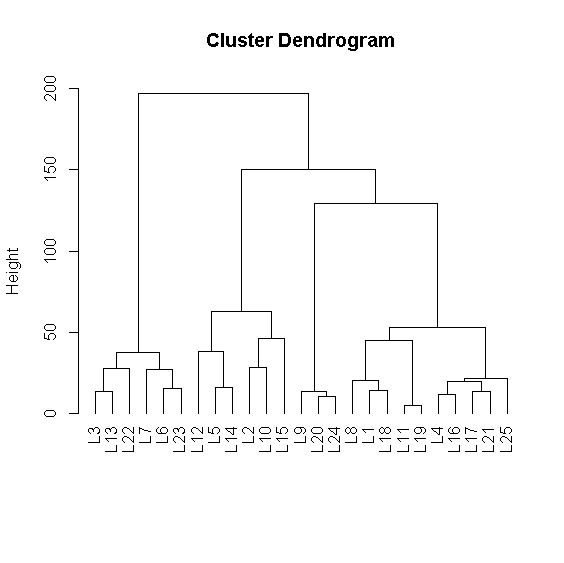
Now we are ready for analysis. First we determine using leaders method 25 leaders. We cluster them further using the hierarchical method and plot the dendrogram.
setwd("C:/Users/Batagelj/work/clamix/clamix.R")
source("C:\\Users\\Batagelj\\work\\clamix\\clamix.R\\clamix3.R")
load("./sr25/sr25.so")
load("./sr25/sr25.meta")
alpha <-rep(1/nVar,nVar)
rez <- leaderSO(SOs,25)
save(rez,file="./sr25/sr25.rez")
hc <- hclustSO(rez$leaders)
plot(hc,hang=-1)
long[rez$clust==9]
long[rez$clust==1]
> save(nVar,nVarP,so,numSO,SOs,file="./sr25/sr25.so") > date() [1] "Sat Nov 03 07:11:15 2012" > alpha <-rep(1/nVar,nVar) > rez <- leaderSO(SOs,25) Step 1 clust 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 130 1075 602 96 375 882 379 28 501 237 168 857 334 175 272 179 17 18 19 20 21 22 23 24 25 581 137 78 88 277 143 165 269 166 [1] 0.4424408 0.4596538 0.4499387 0.4489372 0.4470013 0.4753601 0.4577974 [8] 0.4397338 0.4697205 0.4506480 0.4472091 0.4817797 0.4512397 0.4443368 [15] 0.4435127 0.4418945 0.4802104 0.4537058 0.4458928 0.4456002 0.4613981 [22] 0.4412230 0.4523715 0.4399890 0.4569876 [1] -0.4424408 -0.4596538 -0.4499387 -0.4489372 -0.4470013 -0.4753601 [7] -0.4577974 -0.4397338 -0.4697205 -0.4506480 -0.4472091 -0.4817797 [13] -0.4512397 -0.4443368 -0.4435127 -0.4418945 -0.4802104 -0.4537058 [19] -0.4458928 -0.4456002 -0.4613981 -0.4412230 -0.4523715 -0.4399890 [25] -0.4569876 [1] 54.07908 450.00058 252.98951 39.45177 155.26080 362.11904 156.06820 [8] 11.64665 209.99214 99.61139 69.49693 359.51359 139.47882 73.29974 [15] 113.63847 73.40548 242.24122 57.69107 32.77622 36.53021 116.37494 [22] 59.30112 68.02095 110.01321 69.56174 [1] 3412.563 Times repeat = 10 Step 2 clust 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 234 992 510 161 222 762 309 86 601 232 237 837 173 364 224 153 198 201 140 90 21 22 23 24 25 461 320 259 115 313 ............................................... [22] 136.36349 93.83314 69.10369 96.68143 [1] 2477.077 Step 69 clust 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 330 600 266 286 203 500 430 381 357 459 179 771 324 356 136 216 149 331 101 147 21 22 23 24 25 268 462 329 286 327 [1] 0.4404389 0.4563706 0.4399778 0.4475609 0.4636173 0.4234320 0.4580795 [8] 0.4547127 0.3978984 0.4588445 0.3687990 0.4686583 0.4622807 0.4559260 [15] 0.4426157 0.4374157 0.4350053 0.4663559 0.3511556 0.4381077 0.4347615 [22] 0.4146484 0.4320133 0.4280194 0.4651733 [1] 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 [1] 109.17241 157.86730 89.48315 98.65276 59.81671 141.87021 126.43962 [8] 134.96135 71.08819 145.83863 36.53670 265.04374 120.18657 122.27654 [15] 20.18357 62.41475 47.59801 126.56725 20.70570 43.90852 80.48392 [22] 136.36349 93.83314 69.10369 96.68143 [1] 2477.077 Times repeat = 0 > save(rez,file="./sr25/sr25.rez") > hc <- hclustSO(rez$leaders) > plot(hc,hang=-1) > long[rez$clust==19] [1] "ARCHWAY HOME STYLE COOKIES,SUGAR FREE OATMEAL" [2] "ARCHWAY Home Style Cookies, Chocolate Chip Ice Box" [3] "ARCHWAY Home Style Cookies, Date Filled Oatmeal" [4] "ARCHWAY Home Style Cookies, Dutch Cocoa" [5] "ARCHWAY HOME STYLE COOKIES,FROSTY LEMON" [6] "ARCHWAY Home Style Cookies, Iced Molasses" [7] "ARCHWAY Home Style Cookies, Iced Oatmeal" [8] "ARCHWAY Home Style Cookies, Molasses" ....... [93] "AUSTIN,CHEDDAR CHS ON WAFER CRACKERS,SANDWICH-TYPE" [94] "KEEBLER,CHS & PNUT BUTTER SNDWCH CRACKERS" [95] "KEEBLER,SUGAR CONES" [96] "MURRAY,HONEY GRAHAM" [97] "SUNSHINE,CHEEZ-IT,ORIGINAL CRACKERS" [98] "SUNSHINE,CHEEZ-IT,RED FAT CRACKERS" [99] "SUNSHINE,KRISPY,SOUP & OYSTER CRACKERS (LARGE)" [100] "KEEBLER,ZESTA,SALTINES,ORIGINAL" [101] "KEEBLER,ZESTA,SALTINES W/ WHL WHEAT" >