Blockmodeling
Indirect approach
We read a network in Pajek. The clustering procedure is quite time (and space) consuming for larger networks. Because of this, for clustering the network, we have first to specify the set of nodes to be clustered by providing a corresponding cluster. In our case we want to cluster a whole network
Cluster/Create complete cluster
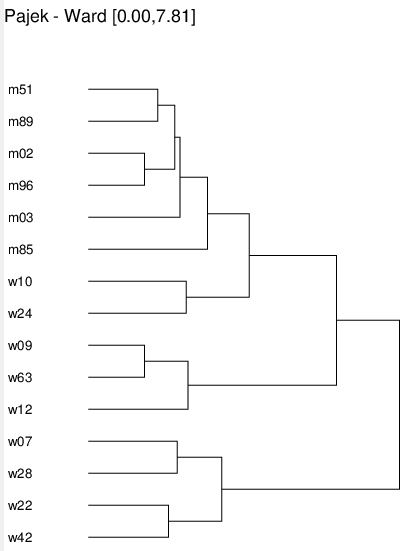
Now we can compute a dissimilarity (Pajek manual, page 51). We select the corrected Euclidean distance. The dissimilarity command automatically makes clustering and ask for the name of EPS file on which it stores the picture of a dendrogram.
Operations/Network+Cluster/Dissimilarity* [5][1][dendro.eps]
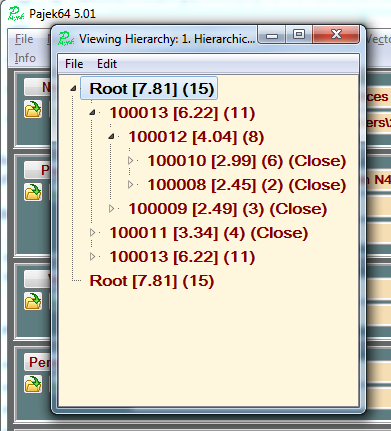
Assume that from the dendrogram we decide to produce 4 clusters. To get them we click on the hierarchy register. A new window opens with a tree display of the hierarchy. We drill into the corresponding levels and close the corresponding tree nodes using Edit/Change Type.
We close the window. Now we can get the partition and, for later use, also a permutation:
Hierarchy/Make partition Hierarchy/Make permutation
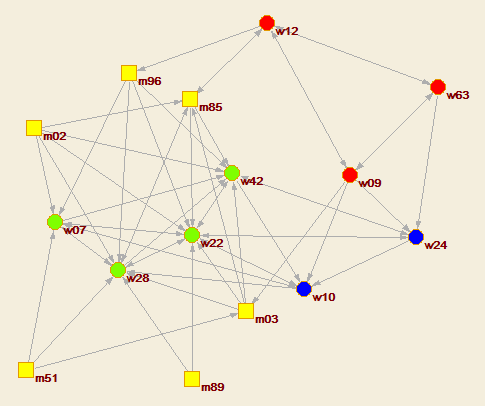
To draw a network with the obtained partiton we use
select the original network Draw/Network+First partition
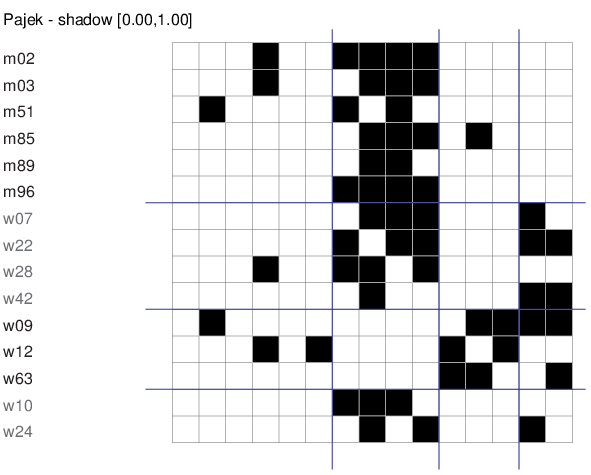
We produce a matrix representation of a clustered network using
File/Network/Export as matrix to EPS/Using Partition+Permutation
Direct approach
Class
read class.net Network/Create Partition/Blockmodeling*/Random start [structural equivalence, 4 clusters] Network/Create Partition/Blockmodeling*/Random start [regular equivalence, 4 clusters]
Tina / LJ student government
read tina.net Network/Create Partition/Blockmodeling*/Random start [regular equivalence, 4 clusters] Network/Create Partition/Blockmodeling*/Random start [user defined, 4 clusters]
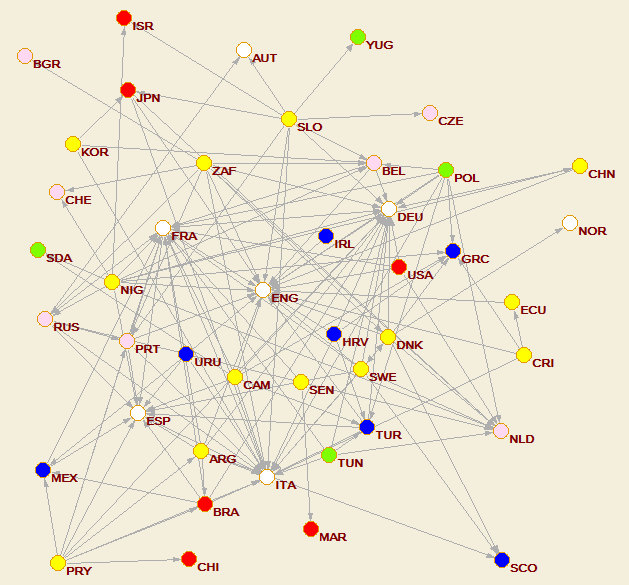
Football market
C:\Users\batagelj\Documents\papers\2017\Moscow\bm
read Football.net Network/Create new network/Transform/Line values/Set all line values to 1 Network/Create Partition/Blockmodeling*/Random start
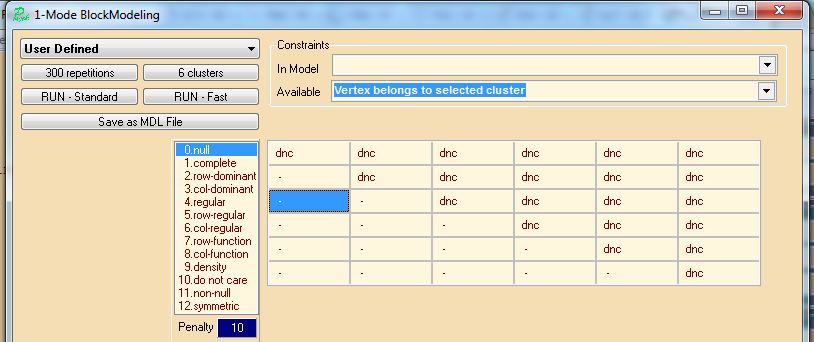
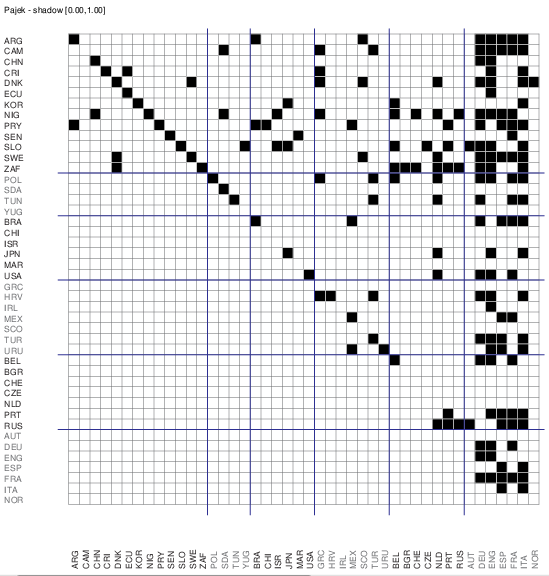
We would like to detect a hierarchical (acyclic) structure in this network. Based on the theorem that the nodes of an acyclic network can be reorder in such a way that the corresponding matrix has a zero lower triangle we can construct a pre-specified blockmodel imposing this restriction. Using the penalty value for a block we can make some errors more important. We assumed a clustering in k=6 clusters.
For a later use we saved the pre-specified blockmodel to the file acyclicDNC6.mdl. We obtained 10 nonequivalent solutions with the criterion function value = 0. Inspecting them in Pajek we selected the following solution because it has well balanced sizes of clusters
We draw the corresponding matrix representation.
select partition Partition/Make permutation File/Network/Export as matrix to EPS/Using Partition+Permutation
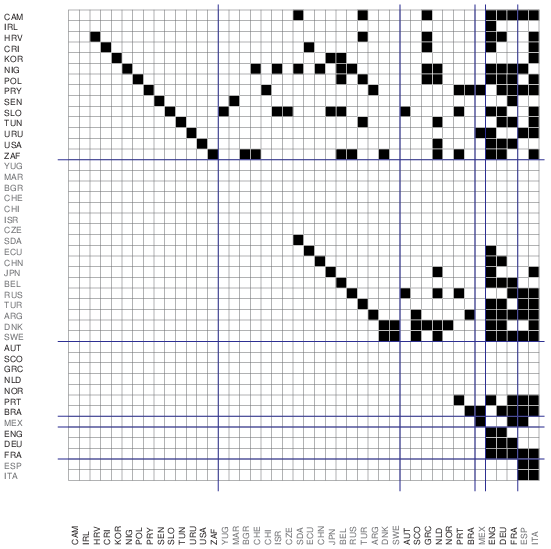
If we assume that the data a correct (no measurement errors) we can determine the acyclic structure faster and exactly using the theorem: if we shrink each strong component of a directed graph into its own model node then the obtained blockmodel is acyclic (with loops allowed). This blockmodel has the smallest number of levels and each node is at its lowest level. Other solutions can be obtained by promoting some nodes to higher level. This procedure can be used also for very large networks.
Network/Create Partition/Components/Strong [1] Operations/Network+Partition/Shrink Network [1][0] Network/Create New Network/Transform/Remove/Loops [yes] Network/Acyclic Network/Depth Partition/Acyclic select strong components partition as First select depth partition as Second Partitions/Functional Composition First*Second Partition/Make Permutation select original network File/Network/Export Matrix to EPS/Using Partition+Permutation
It turns out the our network has 44 nodes and 40 strong components - it is almost acyclic. It requires 6 levels (clusters). The final picture was obtained by manual transposition of some nodes inside clusters.