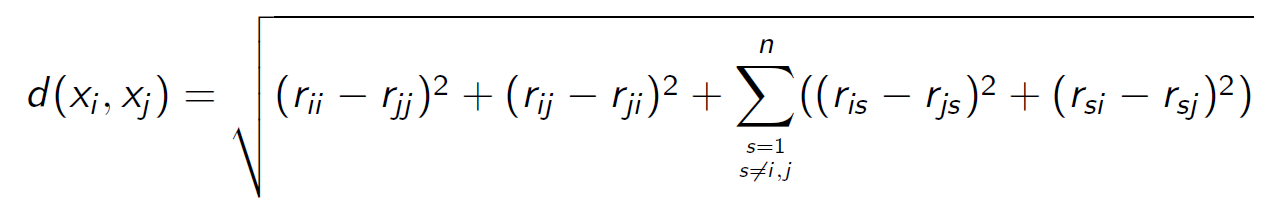Notes
Entering Pajek file
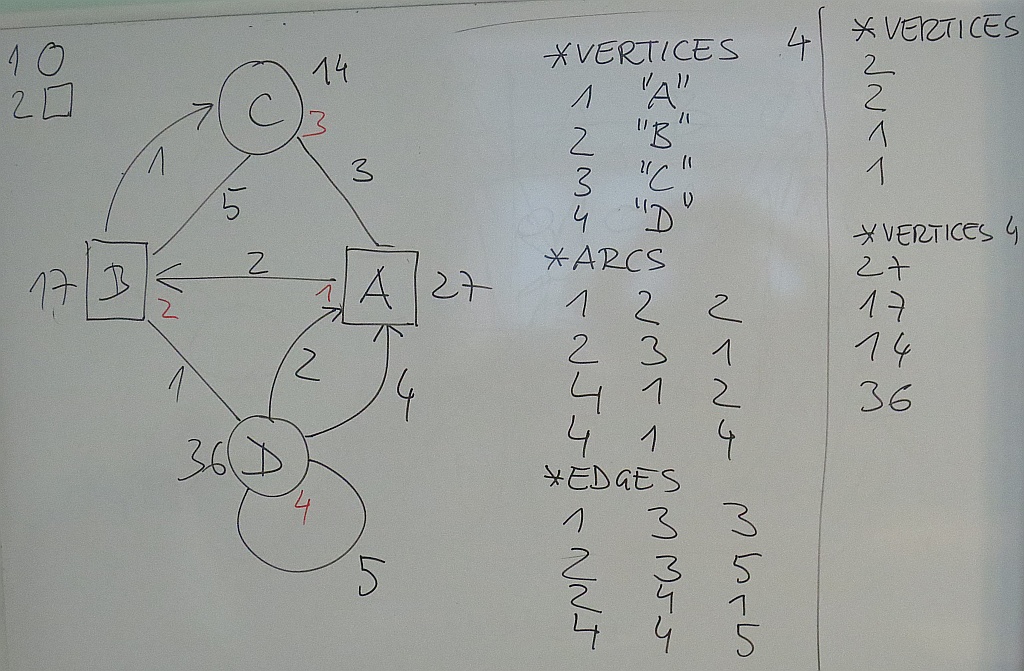
We describe the network from the picture with three files:
ExNet.net
% Example network - ISS7 % Moscow, June 2017 *vertices 4 1 "A" 2 "B" 3 "C" 4 "D" *arcs 1 2 2 2 3 1 4 1 2 4 1 4 *edges 1 3 3 2 3 5 2 4 1 4 4 5
shape.clu
% 1 - circle 2 - square *vertices 4 2 2 1 1
and value.vec
*vertices 4 27 17 14 36
We can combine them into a single file
ExNet.paj
% Example network - 7ISS % Moscow, June 2017 *Network exNet.net *Vertices 4 1 "A" 0.6472 0.4527 0.5000 2 "B" 0.2408 0.5353 0.5000 3 "C" 0.4770 0.7659 0.5000 4 "D" 0.4678 0.1910 0.5000 *Arcs 1 2 2 2 3 1 4 1 2 4 1 4 *Edges 1 3 3 2 3 5 2 4 1 4 4 5 *Partition shape.clu % 1 - circle 2 - square *Vertices 4 2 2 1 1 *Vector value.vec *Vertices 4 27 17 14 36
A file with alternative node names
ExNet.nam
% Example network names - 7ISS % Moscow, June 2017 *vertices 4 1 "Владо" 2 "Борис" 3 "Валя" 4 "Дарья"
has to be saved in Unicode UTF-8 with signiture (BOM). It can be used in Pajek to rename the nodes:
select the network Network/Create New Network/Transform/Add/Vertex Labels/Default [No] Network/Create New Network/Transform/Add/Vertex Labels/From File(s) [ExNet.nam]
Pajek data sets
Transforming migration matrix into Pajek network
https://www.imi.ox.ac.uk/data/demig-data/demig-c2c-data/download-the-data/demig-c2c-data-downloads
Read in Excel and save it in CSV as “bilateralmigrationmatrix20130.csv”.
Move last lines to the front.
Replace ”,” with ””.
Save as “migration2013.csv”
> setwd("C:/Users/batagelj/Downloads/data/migration")
> D <- read.csv2("migration2013.csv",row.names=1,skip=3)
> A <- as.matrix(D)
> dim(A)
[1] 218 218
> A[1:10,1:10]
Afghanistan Albania Algeria American.Samoa Andorra Angola Antigua.and.Barbuda Argentina Armenia
Afghanistan 0 0 0 0 0 0 0 9 0
Albania 0 0 0 0 0 0 0 77 0
Algeria 0 0 0 0 0 0 0 210 0
American Samoa 0 0 0 0 0 0 0 0 0
Andorra 0 0 0 0 0 0 0 47 0
Angola 0 0 0 0 0 0 0 81 0
Antigua and Barbuda 0 0 0 0 0 0 0 0 0
Argentina 0 0 0 0 708 0 0 0 0
Armenia 0 0 0 0 0 0 0 939 0
Aruba 0 0 0 0 0 0 5 0 0
Aruba
Afghanistan 0
Albania 4
Algeria 3
American Samoa 0
Andorra 0
Angola 0
Antigua and Barbuda 5
Argentina 71
Armenia 0
Aruba 0
> A[214:218,214:218]
Zimbabwe Other.North Other.South World X
Zimbabwe 0 1 2 973247 NA
Other North 13164 38330 1168 2713351 NA
Other South 26920 31434 2895 4946635 NA
World 360992 470548 95518 247245059 NA
NA NA NA NA NA
> A <- A[1:217,1:217]
> W <- A[1:216,1:216]
> A[210:216,210:216]
Virgin.Islands..U.S.. West.Bank.and.Gaza Yemen.Rep. Zambia Zimbabwe Other.North Other.South
Virgin Islands (U.S.) 0 0 0 0 0 28 529
West Bank and Gaza 0 0 3740 0 0 0 0
Yemen Rep. 0 1473 0 0 0 0 0
Zambia 0 0 0 0 26909 11 10
Zimbabwe 0 0 0 5149 0 1 2
Other North 2886 2015 2483 861 13164 38330 1168
Other South 2692 20874 34508 6777 26920 31434 2895
>
> n <- nrow(A)
> net <- file("migration2013.net","w"); cat('*vertices ',n,'\n',file=net)
> for(v in 1:n) cat(v,' "',row.names(A)[v],'"\n',sep='',file=net)
> cat('*arcs\n',file=net)
> for(v in 1:n) {
+ for(u in 1:n) if(A[v,u]>0) cat(v,' ',u,' ',A[v,u],'\n',sep='',file=net)
+ }
> close(net)
Pathfinder
Pathfinder determines a skeleton of a weighted network. The weights should be dissimilarities. A migration flow is a similarity s. A simple way to transform it into a dissimilarity d is d = 1/s.
read migration2013.net Network/Create new network/Transform/Line values/Power [-1] Network/Create new network/Transform/Reduction/Pathfinder* [0]
We obtain a simplfied network. We draw it using some automatic procedure (Kamada-Kawai/Free) and manually improve the picture.
pfmig.zip in PF1.net the nodes Other South and Other Nord are removed.
Population size
http://www.prb.org/pdf13/2013-population-data-sheet_eng.pdf
http://www.photius.com/rankings/population/population_2013_0.html
https://www.cia.gov/library/publications/the-world-factbook/rankorder/2119rank.html
We download the file https://www.cia.gov/library/publications/the-world-factbook/rankorder/rawdata_2119.txt and transform it into CSV format: add header, remove commas from numbers, add a separator ; between columns. We save it as popCnt.csv .
> P <- read.csv2("popCnt.csv",row.names=2,strip.white=TRUE)
Fusing the data
> Pnames <- tolower(rownames(P)) > head(Pnames) [1] "china" "india" "european union" "united states" [5] "indonesia" "brazil" > Anames <- tolower(rownames(A)) > head(Anames) [1] "afghanistan" "albania" "algeria" "american samoa" [5] "andorra" "angola" > p <- match(Anames,Pnames) > q <- match(Pnames,Anames) > cbind(which(is.na(p)),Anames[is.na(p)]) > cbind(which(is.na(q)),Pnames[is.na(q)])
Manually find the matchings:
> cbind(which(is.na(p)),Anames[is.na(p)])
[,1] [,2]
[1,] "14" "bahamas the"
[26,] "180" "bahamas, the"
[2,] "28" "brunei darussalam"
[25,] "175" "brunei"
[3,] "39" "channel islands"
[31,] "197" "jersey"
[32,] "205" "guernsey"
[4,] "44" "congo dem. rep."
[5,] "18" "congo, democratic republic of the"
[5,] "45" "congo rep."
[18,] "125" "congo, republic of the"
[6,] "52" "czech republic"
[13,] "87" "czechia"
[7,] "58" "egypt arab rep."
[3,] "16" "egypt"
[8,] "64" "faeroe islands"
[34,] "212" "faroe islands"
[9,] "70" "gambia the"
[21,] "147" "gambia, the"
[10,] "84" "hong kong sar china"
[14,] "101" "hong kong"
[11,] "89" "iran islamic rep."
[4,] "17" "iran"
[12,] "101" "korea dem. rep."
[10,] "51" "korea, north"
[13,] "102" "korea rep."
[7,] "28" "korea, south"
[14,] "105" "kyrgyz republic"
[16,] "115" "kyrgyzstan"
[15,] "106" "lao pdr"
[15,] "104" "laos"
[16,] "115" "macao sar china"
[23,] "170" "macau"
[17,] "116" "macedonia fyr"
[20,] "146" "macedonia"
[18,] "127" "micronesia fed. sts."
[28,] "194" "micronesia, federated states of"
[19,] "134" "myanmar"
[6,] "25" "burma"
[20,] "158" "russian federation"
[2,] "10" "russia"
[21,] "169" "sint maarten (dutch part)"
[35,] "213" "sint maarten"
[22,] "170" "slovak republic"
[17,] "119" "slovakia"
[23,] "178" "st. kitts and nevis"
[33,] "210" "saint kitts and nevis"
[24,] "179" "st. lucia"
[27,] "187" "saint lucia"
[25,] "180" "st. martin (french part)"
[37,] "217" "saint martin"
[26,] "181" "st. vincent and the grenadines"
[30,] "196" "saint vincent and the grenadines"
[27,] "187" "syrian arab republic"
[12,] "66" "syria"
[28,] "208" "venezuela rb"
[8,] "43" "venezuela"
[29,] "210" "virgin islands (u.s.)"
[29,] "195" "virgin islands"
[30,] "211" "west bank and gaza"
[19,] "142" "west bank"
[22,] "153" "gaza strip"
[31,] "212" "yemen rep."
[9,] "48" "yemen"
[32,] "215" "other north"
[33,] "216" "other south"
> cbind(which(is.na(q)),Pnames[is.na(q)])
[,1] [,2]
[1,] "3" "european union"
[11,] "55" "taiwan"
[24,] "171" "western sahara"
[36,] "215" "british virgin islands"
[38,] "219" "gibraltar"
[39,] "221" "anguilla"
[40,] "222" "wallis and futuna"
[41,] "224" "nauru"
[42,] "225" "cook islands"
[43,] "226" "saint helena, ascension, and tristan da cunha"
[44,] "227" "saint barthelemy"
[45,] "228" "saint pierre and miquelon"
[46,] "229" "montserrat"
[47,] "230" "falkland islands (islas malvinas)"
[48,] "231" "norfolk island"
[49,] "232" "christmas island"
[50,] "233" "svalbard"
[51,] "234" "tokelau"
[52,] "235" "niue"
[53,] "236" "holy see (vatican city)"
[54,] "237" "cocos (keeling) islands"
[55,] "238" "pitcairn islands"
and construct the population number vector
> pNA <- c(
+ 14, 28, 39, 44, 45, 52, 58, 64, 70, 84,
+ 89, 101, 102, 105, 106, 115, 116, 127, 134, 158,
+ 169, 170, 178, 179, 180, 181, 187, 208, 210, 211,
+ 212 )
>
> qNA <- c(
+ 180, 175, 197, 18, 125, 87, 16, 212, 147, 101,
+ 17, 51, 28, 115, 104, 170, 146, 194, 25, 10,
+ 213, 119, 210, 187, 217, 196, 66, 43, 195, 142,
+ 48 )
> popP <- P$pop
> head(popP)
[1] 1373541278 1266883598 513949445 323995528 258316051 205823665
> pn <- p
> pn[pNA] <- qNA
> Anames[is.na(pn)]
[1] "other north" "other south"
> popP[142] <- popP[142]+popP[153]
> popP[197] <- popP[197]+popP[205]
> pop <- popP[pn]
> Anames[is.na(pop)]
[1] "other north" "other south"
> n <- nrow(A)-2
> net <- file("migration2013B.net","w"); cat('*vertices ',n,'\n',file=net)
> vec <- file("migration2013pop.vec","w"); cat('*vertices ',n,'\n',file=vec)
> for(v in 1:n) cat(v,' "',row.names(A)[v],'"\n',sep='',file=net)
> cat('*arcs\n',file=net)
> for(v in 1:n) {
+ cat(pop[v],'\n',file=vec)
+ for(u in 1:n) if(A[v,u]>0) cat(v,' ',u,' ',A[v,u],'\n',sep='',file=net)
+ }
> close(net); close(vec)
> names(pop) <- Anames
> save(A,pop,file="migration2013.RData")
Clustering the migration network
To make countries (described by rows in migration matrix) comparable we have to normalize them. There are at least two options:
- divide each row by the sum of it entries: conditional probability that a migrant from the first country will migrate to the second country;
- divide each row by the size of a country population: probability that a citizen of the first country will migrate to the second country.
The function netDist(A) implements the corrected Euclidean dissimilarity between rows (but not columns!!!) of a matrix A.
> S <- apply(A,1,sum)
> T <- A/S
> netDist <- function(A){ n <- nrow(A)
+ D <- matrix(nrow=n,ncol=n,dimnames=dimnames(A)); diag(D) <- 0
+ for(v in 2:n){
+ for(u in 1:(v-1)) {
+ d <- sum((A[v,]-A[u,])**2) - (A[v,u]-A[u,u])**2 - (A[v,v]-A[u,v])**2 +
+ (A[v,u]-A[u,v])**2 + (A[v,v]-A[u,u])**2
+ D[v,u] <- D[u,v] <- sqrt(d)
+ }
+ }
+ return(as.dist(D))
+ }
> D <- netDist(T)
> r <- hclust(D,method="ward.D")
> plot(r,hang=-1,cex=0.2,main="Migrations 2013, profiles")
> head(Anames[r$order],n=10)
[1] "belize" "guatemala"
[3] "cayman islands" "el salvador"
[5] "marshall islands" "mexico"
[7] "puerto rico" "palau"
[9] "micronesia fed. sts." "northern mariana islands"
> per <- file("migration2013ward.per","w"); cat('*vertices ',n,'\n',file=per)
> for(v in 1:n) cat(r$order[v],'\n',file=per); close(per)
>
> n <- n-2
> Ap <- A[1:n,1:n]
> po <- pop[1:n]
> N <- Ap/po
> D <- netDist(N)
> r <- hclust(D,method="ward.D")
> plot(r,hang=-1,cex=0.2,main="Migrations 2013, intense")
> Nt <- t(N)
> D <- netDist(Nt)
> r <- hclust(D,method="ward.D")
> plot(r,hang=-1,cex=0.2,lwd=0.5,main="Migrations 2013, intense/transpose")
> B <- Ap
> B[B>0] <- 1
> D <- netDist(B)
> r <- hclust(D,method="ward.D")
> plot(r,hang=-1,cex=0.2,lwd=0.5,main="Migrations 2013, binary")
> Bt <- t(B)
> D <- netDist(Bt)
> r <- hclust(D,method="ward.D")
> plot(r,hang=-1,cex=0.2,lwd=0.5,main="Migrations 2013, binary/transpose")